DNA Profiling – 70 course points
The purpose of this assignment is to practice using Abstract Data Types to analyze DNA.
Refer to our Programming Assignments FAQ for instructions on how to install VScode, how to use the command line and how to submit your assignments.
Programming
The assignment has two components:
- Coding (67 points) submitted through Autolab.
- Reflection (3 points) submitted through a form.
- Submit the reflection AFTER you have completed the coding component.
- Be sure to sign in with your RU credentials! (netid@scarletmail.rutgers.edu)
- You cannot resubmit reflections but you can edit your responses before the deadline by clicking the Google Form link, signing in with their netid, and selecting “Edit your response”
We provide a zip file in Autolab containing DNA.java and input files.
Watch this video that shows how to open the folder in VSCode.
Observe the following rules:
- DO NOT add any import statements
- DO NOT add the project statement
- DO NOT change the class name
- DO NOT change the headers of ANY of the given methods
- DO NOT add any new class fields
- DO NOT use System.exit()
DNA Overview
DNA is the carrier of genetic information that all living things are made of. We can use DNA to tell us useful information, such as finding to whom it belongs and their relatives. So, DNA profiling can take a look at DNA to identify to whom it belongs.
- DNA is a sequence of protein molecules called nucleotides, arranged into a particular shape (a double helix). Each nucleotide of DNA contains one of four different bases: adenine (A), cytosine (C), guanine (G), or thymine (T). Every human cell has billions of these nucleotides arranged in sequence (genome).
How can this identification be accomplished? DNA is composed of genes (which all humans share) and “junk” DNA (sections of non-coding DNA in between the genes that are highly variable between individuals). By looking at these sections of “junk DNA” we can identify a person through a DNA sample or find relatives.
What are we looking for in this junk DNA? Inside of these sections of the DNA, there is a high variance nucleotide patterns. Due to this high variance we can uniquely identify individuals based on the similarities in the STRs (Short Tandem Repeats) within the junk DNA.
What are STRs and how can I use them to tell me what I want to know? STRs are Short Tandem Repeats, which are short sequences of DNA that repeat back-to-back in DNA sequences numerous times. AGAT and ACGT are examples of short DNA sequences which we can use as STRs. Similarities in STRs would indicate genetic relationships.
Example DNA Sequence: AGATAGATAGATACGTACGT
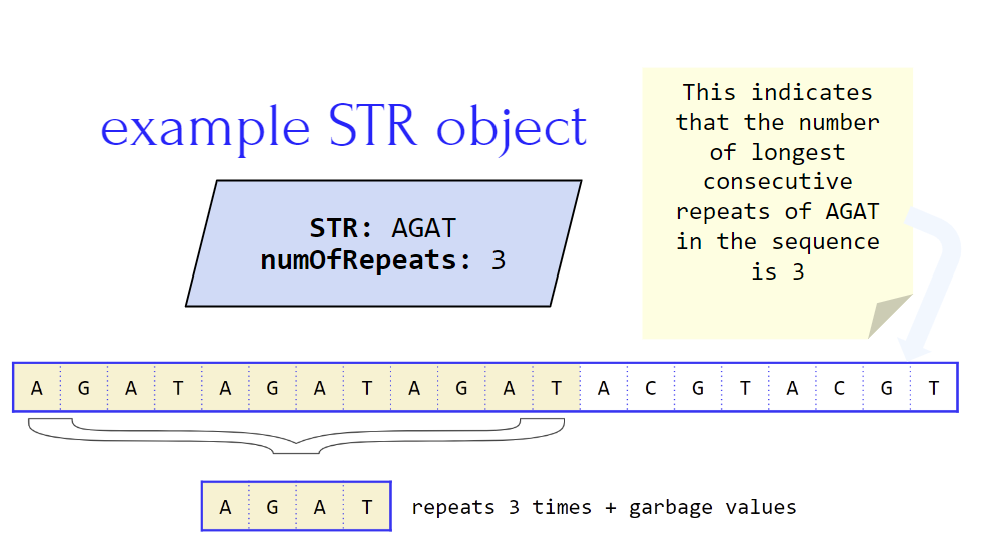
Here, you see that the STR “AGAT” is repeated three times followed by another STR “ACGT” repeated twice (this is just an example of many possibilities). The more STRs we use to analyze a DNA sequence the more accurate our findings are. When looking at STRs in a DNA sequence we want to look at the maximum consecutive times it is repeated within one sequence to find the highest variance of the STR.
The FBI uses 20 different STRs when processing DNA for their Combined DNA Index System (CODIS) database.
- For simplicity, we will be using 2 to 4 STRs that are a few nucleotides long.
The goal of this assignment is to carry out a DNA profiling method called STR analysis, where we look at STRs to:
- Find out who the unknown person is
- Find out who their possible parents could be
Implementation
Overview of Files
DO NOT edit ANY class besides DNA.java
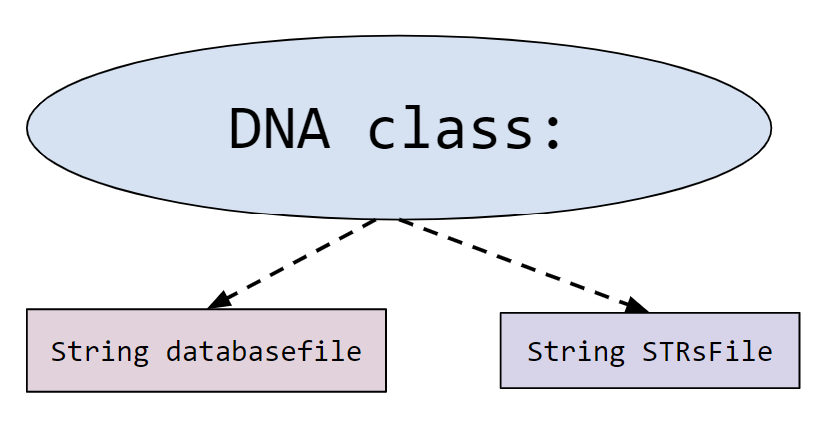
- DNA.java: contains some provided methods in addition to annotated method signatures for all the methods you are expected to fill in. You will write your solutions in this file. This is the only file which will be submitted for grading.
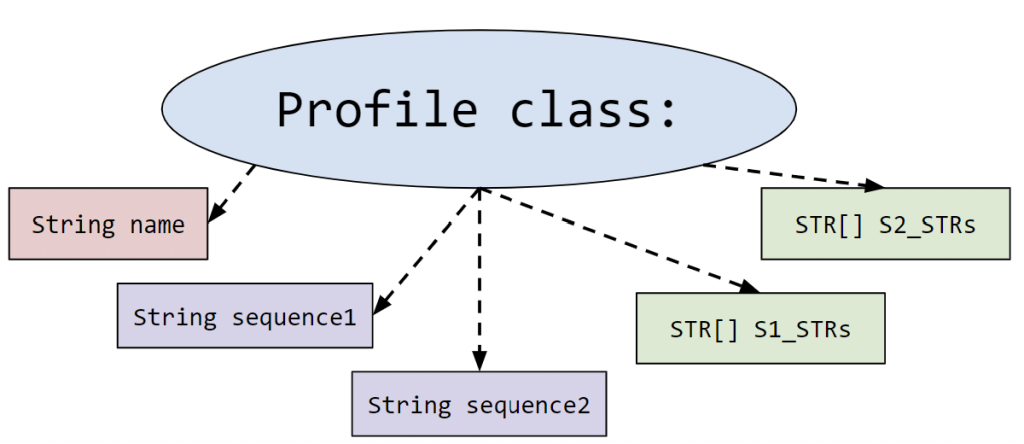
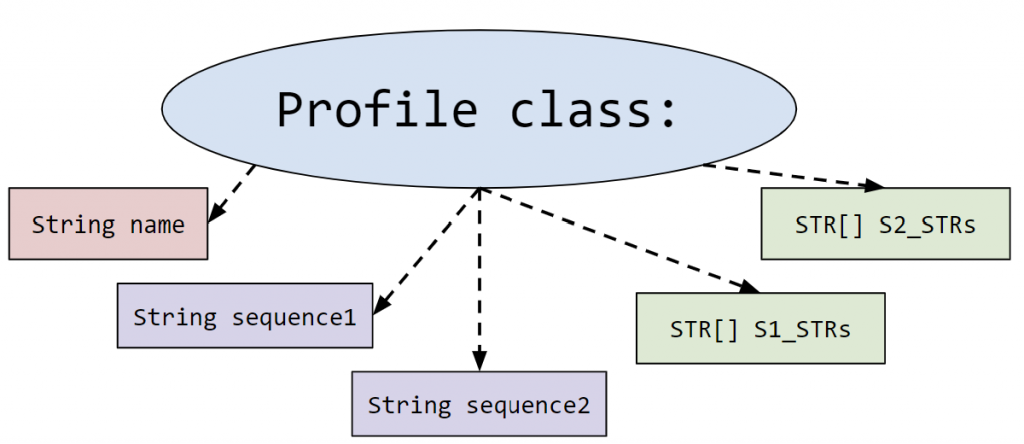
- Profile.java: This represents the profile of a person. More specifically it holds the information about the 23rd pair of chromosomes that is used for identifying parent/offspring relationships.
It contains the person’s name and two DNA sequences (one from each parent). These sequences are read from the input file in createDatabaseOfProfiles().
- STR.java: This class represents the Short Tandem Repeats. An STR is a short sequence of DNA nucleotides that repeat back-to-back numerous times.
- StdIn: is used by the driver.
- StdOut: is used by the driver.
- Driver: you can use this to test any of your methods interactively. Feel free to edit this class, as it is provided only to help you test. It is not submitted and it is not used to grade your code. Scroll down for instructions on how to compile and test your assignment.
- Multiple txt files: files that can be read by the driver as test cases.
- profiles.txt – this file contains the actual DNA sequences of the people. These will be long Strings of A, C, G, and T, which we will scan through for the existence of STRs.
- allSTRs0.txt and allSTRs1.txt – will be used to process the profiles.txt file to make the actual database. allSTRs0.txt has 1 STR, and allSTRs1.txt has 3 STRs. The more STRs there are, the more precise the results will be.
- matching0.txt – will be used to test if an unknown DNA sequence exists in the database
- offspring0.txt – will be used to test if potential parents of an unknown DNA sequence exist in the database.
You are allowed (and encouraged) to make your own test cases to test different edge cases! You may use the data.txt file to make new test cases, or come up with your own version of the profiles.txt file!
DNA.java
This class has two instance variables:
What are instance variables? Variables declared outside the scope of methods but inside the class. Each time an instance of the class is created a new instance variable is created.
- database holds the profiles of all people in the database.
- STRsOfInterested holds the STRs (Short TAndem Repeats) we are interested in looking for while DNA profiling.
DNA.java
This class has two instance variables:
- database holds the profiles of all people in the database.
- STRsOfInterest holds the STRs (Short Tandem Repeats) we are interested in looking for while DNA profiling.
Methods implemented by you:
For all of these, look at the comments above the methods for more information.
This method is already written for you. DO NOT EDIT THIS METHOD. This method creates a DNA object and calls createDatabaseOfProfiles and readSTRsOfInterest.
1. createDatabaseOfProfiles (String filename)
Input parameter: profile files
Input file format:
- 1 line containing an integer, p, with the number of profiles/people in the file
- for each p profiles in the file
- 1 line containing the person’s name
- 1 line containing the first sequence of STRs that comes from one parent
- 1 line containing the second sequence of STRs that comes from the other parent
This method creates and populates the database array with p number of profiles from the input file.
Approach:
- Create the database array to hold the profiles (don’t forget about the instance variables)
- Read the number of profiles from the input file
- Read the profile information from the input file
- For each person in the file
- creates a Profile object with the information from file (see input file format below)
- insert the newly created profile into the next position in the database array
💡*Hint: You can use StdIn.String() to read 1 (one) string from the file.
What if we don’t have an attribute at the time of making the object? We set the attribute as NULL.
2. readAllSTRsOfInterest (String filename)
Input parameter: allSTRs files
Input file format:
- 1 line containing an integer, s, with the number of STRs in the file
- for each s in the file
- 1 line containing the STR
This method creates and populates the STRsOfInterest array with s number of STRs from the input file.
Similar approach as createDatabaseOfProfiles
💡*Hint: You can use StdIn.readString() to read 1 (one) string from the file.
How do we get/set each attribute of an object? objectname.getAttribute() or objectname.setAttribute(value) or objectname.attribute = value
After createDatabaseOfProfiles and readAllSTRsOfInterest are completed, you can run the following in the driver.
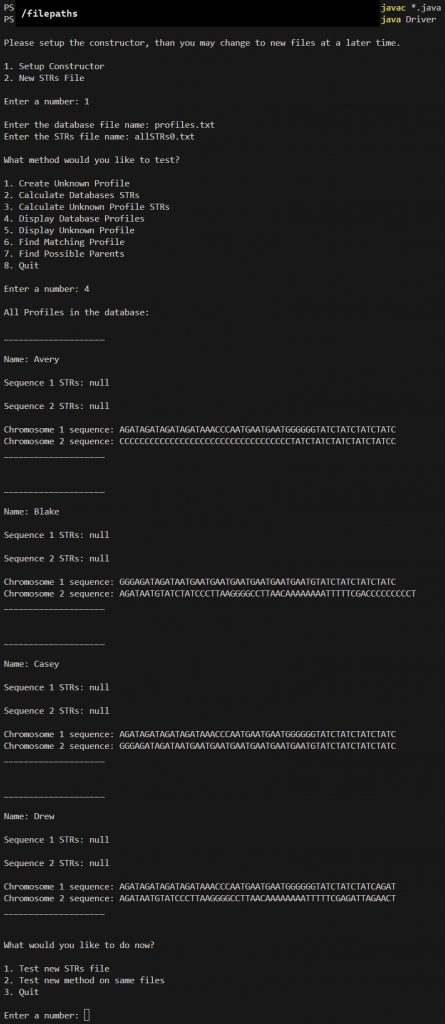
Option 1
profiles.txt
allSTRs0.txt
Option 4
Notice how the STRs are still NULL since we initialized them as NULL in the createDatabaseOfProfiles method.
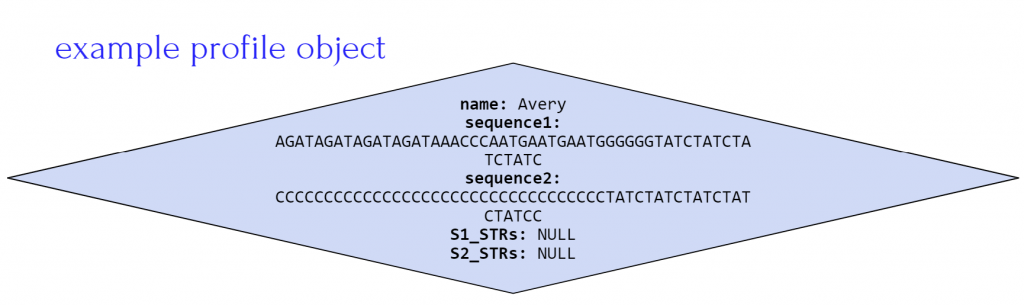
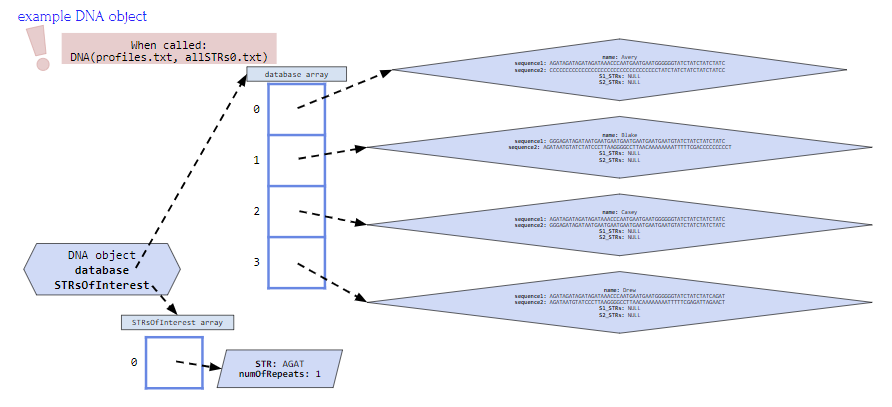
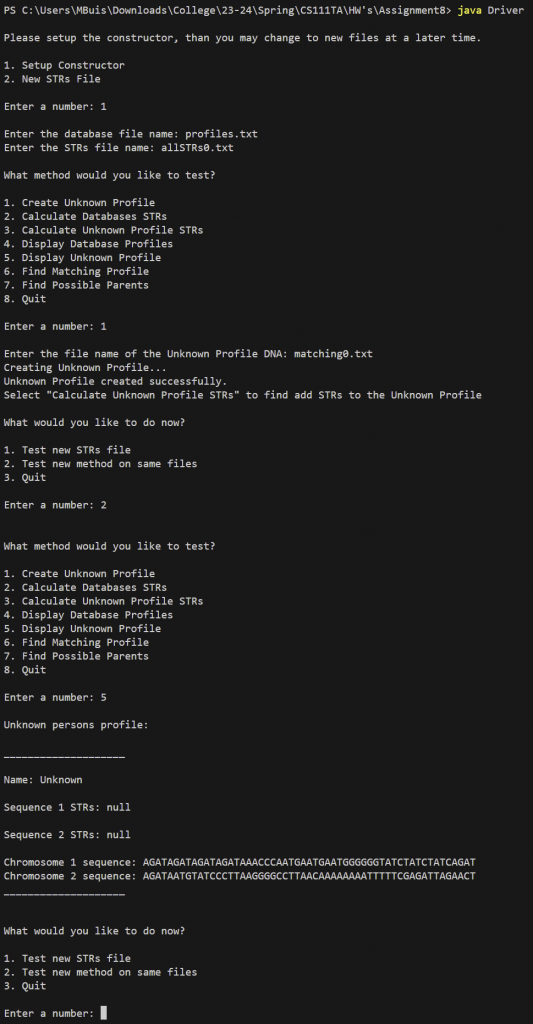
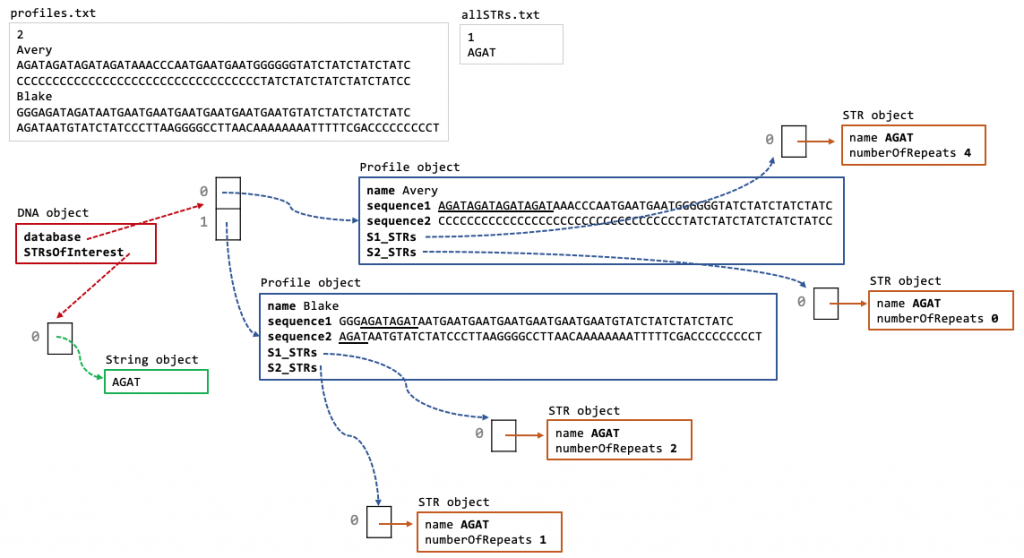
In addition if the constructor is called using DNA(profiles.txt, allSTRs0.txt) the DNA object would look like the figure below where:
- database instance variable array contains four profiles, one for Avery, Blake, Casey and Drew
- STRsOfInterest instance variable array contains one string for STR AGAT
3. createUnkownProfile (String filename)
Input parameter: matching files
Input file format:
- first line containing a DNA sequence
- second line containing a DNA sequence
This method creates and returns the profile for the known DNA sequence from the input file.
Approach:
- Set profile name to “Unknown” because they are currently unknown
- Set the S1_STRs and S2_STRs to NULL
- Set sequence1 to the first line of the file
- Set sequence2 to the second line of the file
- Return the Profile object
💡*Hint: You can use StdIn.readString() to read 1 (one) string from the file.
Option 1
profiles.txt
allSTRs0.txt
Option 1
matching0.txt
Option 2
Option 5
Notice how the STRs are still NULL since we initialized them as NULL.
Notice how their name is “Unknown” since we manually set their name in createUnknownProfile by this method.
Given a DNA sequence and a singular STR, this method finds the longest number of consecutive repeats that STR appears in the sequence and then returns an STR object with:
- the STR name
- longest number of consecutive repeats
Approach:
- Set new STR with parameters STR and 0 as numberOfRepeats
- Check if the STR is longer than the sequence
- Traverse through the sequence checking for repeating STRs
- Return the STR object
*Note: If STR has more characters than the sequence then the return object will have 0 (zero) numberOfRepeats.
💡*Hint: How do I search for the longest number of consecutive STR repeats in the sequence? look at the Java String class and the included methods
5. createProfileSTRs(Profile profile, String[] allSTR)
Input parameter: Profile profile, String[] allSTR
This method takes a profile and a String[] and populates then adds the STRs to the profile. (Remember we set them as NULL before).
Approach:
- Populate some new S1_STRs and S2_STRs arrays by traversing the allSTR[]
- Use setter method to set S1_STR and S2_STR to profile
*Note: You will need to use the findSTRInSequence method you just wrote
💡*Hint: Think about the attributes of STRs
Input parameter: NA
This method creates and updates the STRs for each profile in the database.
Approach:
- Similar strategy used in createDatabaseOfProfiles and readAllSTRsOfInterest methods
*Note: You will need to use the createProfileSTRs that you just wrote, for each profile in the database
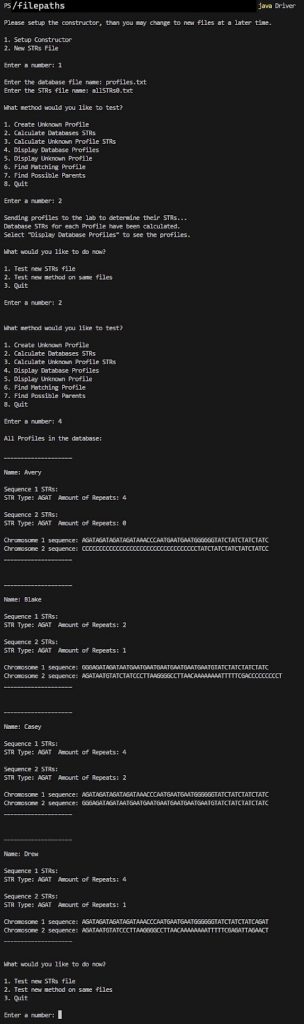
We are now ready to do some analysis! Now that the database is created and the STRs for each Profile have also been created according to the STRsOfInterest!🎉
Option 1
profiles.txt
allSTRs0.txt
Option 2
Option 2
Option 4
Notice how the STRs are now populated from the findSTRInSequence and createProfileSTRs methods.
Option 1
profiles.txt
allSTRs0.txt
Option 1
matching0.txt
Option 2
Option 3
Option 2
Option 5
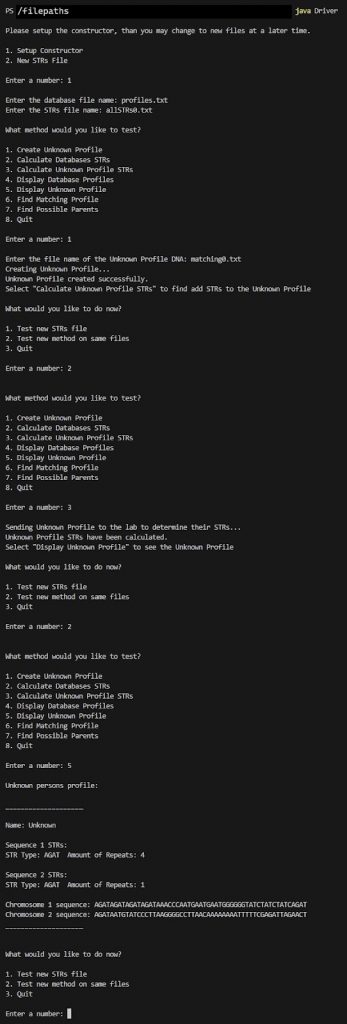
Notice how the STRs are now populated from the findSTRInSequence and createProfileSTRs methods.
7. identicalSTRs (STR[] s1, STR[] s2)
Input parameter: STR[] s1, STR[] s2
This method copmares the two STR parameter arrays to determine if they are identical. Two STRs are identical if the objects at s1[i] and s2[i] contain the same information for every i in the arrays
Approach:
- Check if s1[0] is identical to (matches) s2[0]
- Check if s1[1] is identical to (matches) s2[1] etc etc
*Note: Assume the parameter arrays are the same length
💡*Hint: Think about how you compare objects vs primitives.
Input parameter: STR[] s1, STR[] s2
This method looks through the database searching for profiles whose S1 matches the unknown profile’s unknownProfilesS1_STRs and returns an array list of all the profiles.
Approach:
- Initalize an ArrayList
- For each profile in the database whose sequence1 matches unknownProfilesS1_STRs
- add to ArrayList
- Return the ArrayList
If no match is found the returned ArrayList will be empty.
*Note: Use the identicalSTRs method to compare two profiles.
💡*Hint: Look at the ArrayList methods.
Option 1
profiles.txt
allSTRs0.txt
Option 1
matching0.txt
Option 2
Option 2
Option 2
Option 3
Option 2
Option 6
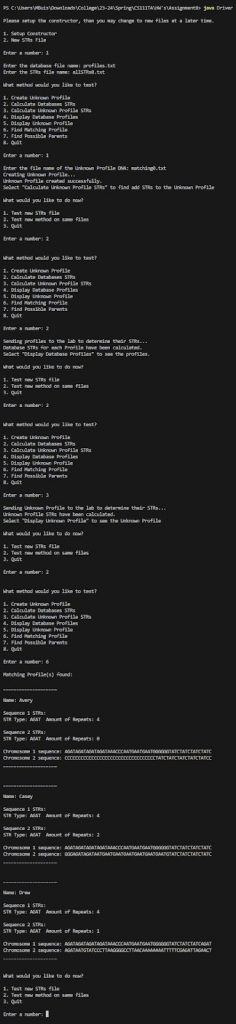
Notice how our profiles are fully updated and we are displaying all profiles that have matching sequence1.
Option 1
profiles.txt
allSTRs1.txt
Option 1
matching0.txt
Option 2
Option 2
Option 2
Option 3
Option 2
Option 6
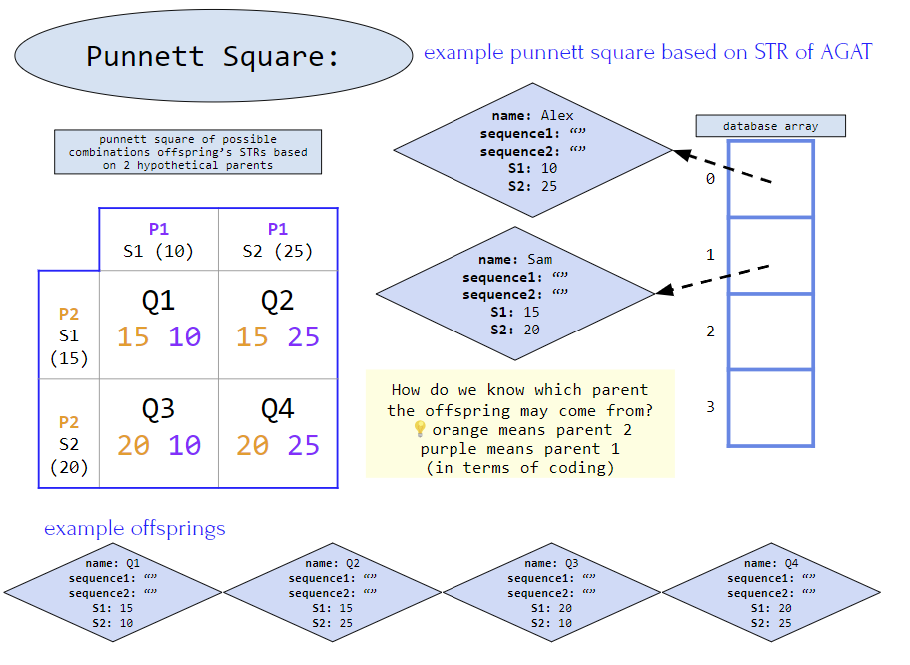
8.5 punnetSquare(STR[] firstParent, STR[] inheritedFromFirstParent,STR[] secondParent, STR[] inheritedFromSecondParent)
This method is already written for you. DO NOT EDIT THIS METHOD. This method takes in 1 STR from each parent and compares it to each STR in the offspring and is called in findPossibleParents.
9. findPossibleParents (STR[] S1_STRs, STR[] S2_STRs)
This method is already written for you, but it DOES NOT work as intended. Your job is to fix this method!
- Check out this video that explains how Punnett squares work and the logic to coding one.
- Some background: Each parent passes down half of their DNA to their offspring, making it possible to trace someone’s parents from their DNA (this is also the basic premise of paternity testing as well). Therefore we look at possible pairing of STRs between parents and see if it matches a possible outcome in the punnett square
Approach:
- First for loop:
- Check the 4 combinations between the database STR# and the offspring STR#
- Second for loops:
- Check each quadrant’s S1 and S2 against the parent STR
Option 1
profiles.txt
allSTRs0.txt
Option 1
offspring0.txt
Option 2
Option 2
Option 2
Option 3
Option 2
Option 7
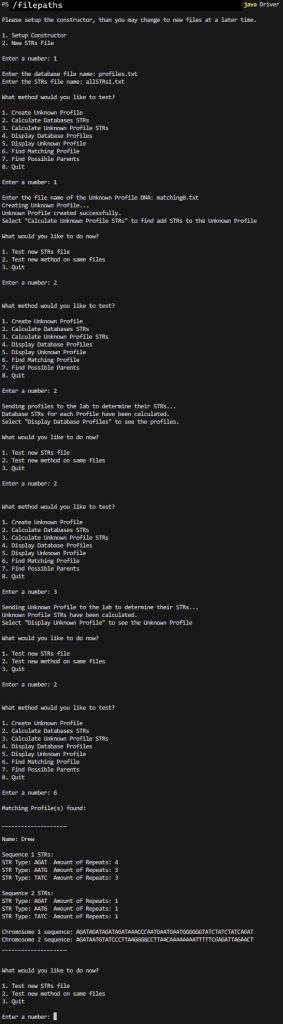
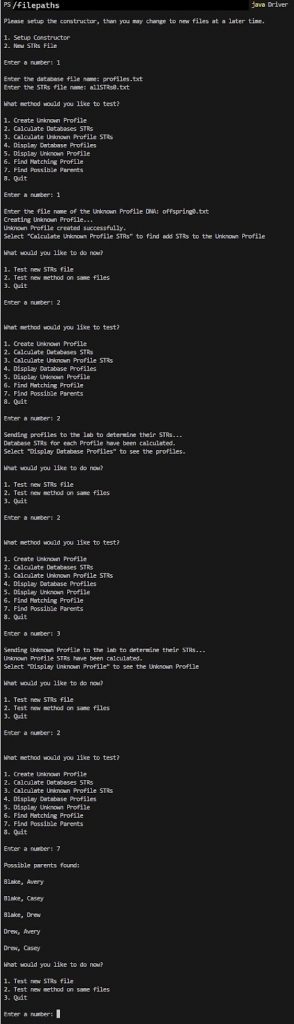
We have returned the profile names of possible parents.
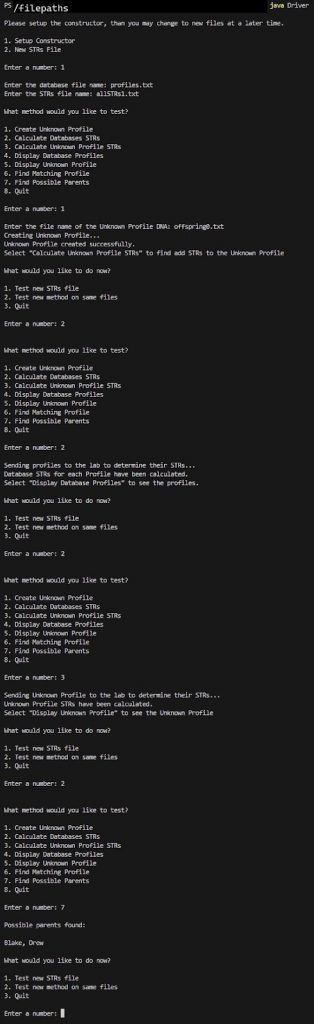
Option 1
profiles.txt
allSTRs1.txt
Option 1
offspring0.txt
Option 2
Option 2
Option 2
Option 3
Option 2
Option 7
We have returned the profile names of possible parents.
** notice how using allSTRs1.txt results in more precise answers?
Executing and Testing
To compile, go to the DNA directory/folder, and run the command javac *.java
To run the compiled Driver, use the command java Driver
What DNA methods need to be completed to run Driver methods strictly without compile errors? (these methods are the bare minimum and will result in some object attributes still being NULL but this will allow you to test the methods as you write them)
What methods need to be completed to run what driver methods? (see below)
- Create Unknown Profile
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- createUnknownProfile
- Display Database Profiles (incomplete at this stage)
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- Display Unknown Profile STRs (complete at this stage)
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- createUnknownProfile
- Calculate Databases AND Display Database Profiles (complete at this stage)
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- findSTRInSequence
- createProfileSTRs
- Calculate Unknown Profile STRs AND Display Unknown Profile STRs (complete at this stage)
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- createUnknownProfile
- findSTRInSequence
- createProfileSTRs
- Find Matching Profile (** guaranteed success if you put in matching.txt files)
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- createUnknownProfile
- findSTRInSequence
- createProfileSTRs
- identicalSTRs
- findMatchingProfiles
- Find Possible Parents (** guaranteed success if you put in offspring.txt files)
- createDatabaseOfProfiles
- readAllSTRsOfInterest
- createUnknownProfile
- findSTRInSequence
- createProfileSTRs
- indenticalSTRs
- findMatchingProfiles
- findPossibleParents
Before submission
Collaboration policy. Read our collaboration policy here.
Submitting the assignment. Submit DNA.java via the web submission system called Autolab. To do this, click the Assignments link from the course website; click the Submit link for that assignment.
Getting help
If anything is unclear, don’t hesitate to drop by office hours or post a question on Piazza.
- Find instructors office hours here
- Find tutors office hours in Canvas -> Tutoring, RU CATS
- Find head TAs office hours here
- POST on Piazza in Canvas -> Piazza
- In addition to office hours we have the CAVE (Collaborative Academic Versatile Environment), a community space staffed with lab assistants which are undergraduate students further along the CS major to answer questions.
Uses libraries and content from Introduction to Computer Science book.
Inspiration from a Nifty assignment by Brian Yu and David J Malan.
Assignment by Seth Kelley and Aastha Gandhi